LipidXplorer Principles
The Workflow of LipidXplorer
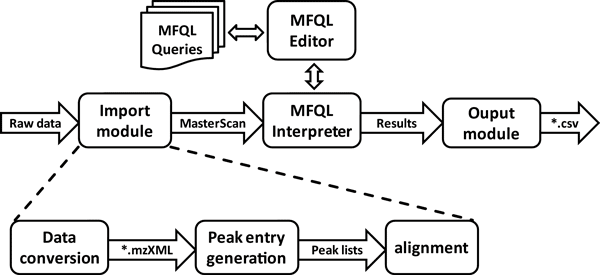
The working of LipidXplorer can be depicted as a simple workflow scheme.
The import module takes the raw mass spectra and converts them into *.mzXML file format (if not given as such). Peaks a generated, aligned and stored in the MasterScan. The MasterScan is basis for LipidXplorers lipid interpretation. The Molecular Fragmentation Query Language (MFQL) Interpreter takes MFQL Queries, which are composted with the MFQL Editor and runs the queries on the MasterScan. The result, which are the identified lipids, are sent to the Ouput module. Here the results are formatted and output as *.csv file.
The MasterScan database
First step in the LipidXplorer workflow is to import all spectra into LipidXplorers own spectra database called MasterScan. The MasterScan contains all information of the whole experiment. This includes all acquisitions of all technical and biological repeats. The spectra are aligned and structured such that all information is easy to access. Such we have an ideal database for a dedicated query language.
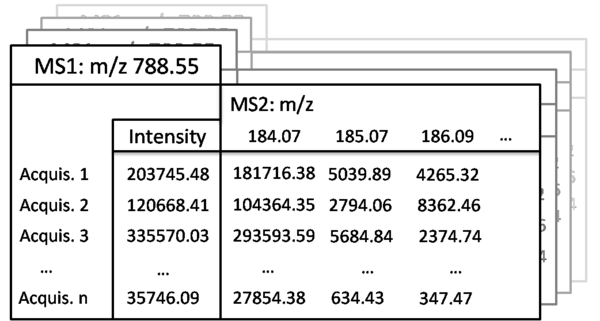
The MasterScan is schematically depicted here as file cabinet addressed on the top-level by the precursor masses (MS1) while intensities are assigned to each acquisition (Acquis.). In this example the precursor mass is m/z 788.55 and n samples are present. If MS2 experiments were acquired a list of fragment masses is linked to their precursor mass and intensities are assigned for each acquisition. In this example MS2 peaks of m/z 184.07, m/z 185.07 and m/z 788.55 are shown with their intensities for each acquisition (Acquis.).
Lipid identification with a query language
Query languages are computer languages used to make queries into databases and information systems (see [Wikipedia: Query language])
To our knowledge, this is the first time a query language is used to identify lipids from mass spectra. Thereby we use the specific fragmentation pathways lipids have in order to identify lipids. The level of accuracy is given only by the limits of the mass spectrometer, but not by the lipid identification software itself.
Using a query language gives great freedom in using different types of mass spectrometer and in how to identify and extract information.