Difference between revisions of "LipidXplorer MFQL"
(→A short tutorial) |
(→Part 2: The IDENTIFY section) |
||
| (53 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | ==Introduction== | + | == Introduction == |
| − | MFQL is the first query language developed for the identification of molecules | + | MFQL is the first query language developed for the identification of molecules in complex shotgun spectra datasets. It formalizes the available or assumed knowledge of lipid fragmentation pathways into queries that are used for probing a MasterScan database. |
| − | in complex shotgun spectra datasets. It formalizes the available or assumed | ||
| − | knowledge of lipid fragmentation pathways into queries that are used for | ||
| − | probing a MasterScan database. | ||
| − | ===Structural complexity of lipid species and sum composition constraints=== | + | === Structural complexity of lipid species and sum composition constraints === |
| − | [[Image:Figure5.png|600px | + | [[Image:Figure5.png|center|600px|Structural complexity of lipid species and sum composition constraints]] '''The figure shows the basic lipid structure and some characteristics specific for lipids using the example of a PC species.''' Let us consider PC as a representative example: PC molecules consist of a posphorylcholine head group attached to the glycerol backbone at the sn-3 position, while fatty acid moieties occupy sn-1 and sn-2 positions (alternatively, a fatty alcohol moiety could be attached at the sn-1 position). Fatty acid moieties differ by the number of carbon atoms and double bonds, but also by the relative location at the glycerol backbone, so that isomeric structures having exactly the same fatty acid moieties are possible. Note that isomeric structures are always isobaric, whereas isobaric molecules are not necessarily isomeric. Most generic constraints ("All lipids of PC class" or "All PC esters") encompass sum compositions of species with common naturally occurring fatty acids. However, because of the fatty acid variability, some species of other lipid classes (such as, PE) might meet the same constraint. Therefore, for most common glycerophospholipid classes, the characterization of individual molecular species could not solely rely on their intact masses, irrespective of how accurately were they measured. MS/MS experiments that produce structure-specific ions contribute more specific constraints, such as the number of carbons and double bonds in individual moieties, characteristic head group fragment, characteristic loss of a fatty acid moiety, among others. Within a MFQL query, these constraints can be bundled by Boolean operations. |
| − | Let us consider PC as a representative example: PC molecules consist of a | ||
| − | posphorylcholine head group attached to the glycerol backbone at the sn-3 | ||
| − | position, while fatty acid moieties occupy sn-1 and sn-2 positions (alternatively, | ||
| − | a fatty alcohol moiety could be attached at the sn-1 position). Fatty acid | ||
| − | moieties differ by the number of carbon atoms and double bonds, but also by | ||
| − | the relative location at the glycerol backbone, so that isomeric structures | ||
| − | having exactly the same fatty acid moieties are possible. Note that isomeric | ||
| − | structures are always isobaric, whereas isobaric molecules are not necessarily | ||
| − | isomeric. Most generic constraints ("All lipids of PC class" or "All PC esters") | ||
| − | encompass sum compositions of species with | ||
| − | However, because of the fatty acid variability, some species of other lipid | ||
| − | classes (such as, PE) might meet the same constraint. Therefore, for most | ||
| − | common glycerophospholipid classes, the characterization of individual | ||
| − | molecular species could not solely rely on their intact masses, irrespective | ||
| − | of how accurately were they measured. MS/MS experiments that produce | ||
| − | structure-specific ions contribute more specific constraints, such as the | ||
| − | number of carbons and double bonds in individual moieties, characteristic | ||
| − | head group fragment, characteristic loss of a fatty acid moiety, among others. | ||
| − | Within a MFQL query, these constraints can be bundled by Boolean operations. | ||
| − | ==A short tutorial== | + | == A short tutorial == |
| − | Below we present an | + | Below we present an example of composing a MFQL query for identifying PC lipids in a typical shotgun dataset. |
| − | example of composing a MFQL query for identifying PC lipids in a typical shotgun dataset. | ||
| − | In MS/MS experiments (see [[# | + | In MS/MS experiments (see [[#MFQL_identification_of_phosphatidylcholines_.28PC.29]]), molecular cations of PC species produce specific phosphorylcholine fragments of their head group having the sum composition of 'C5 H15 O4 N1 P1' and m/z 184.07 (see [[#MFQL_identification_of_phosphatidylcholines_.28PC.29]]). The identification of PC species starts with the identification of probable precursors in the MS spectrum using the accurately determined masses and proceeds with identifying phosphorylcholine headgroup fragment in the MS/MS spectra (see [[#MFQL_identification_of_phosphatidylcholines_.28PC.29]]). |
| − | molecular cations of PC species produce specific phosphorylcholine fragments of | ||
| − | their head group having | ||
| − | the sum composition of 'C5 H15 O4 N1 P1' and m/z 184.07 (see [[# | ||
| − | identification of PC species starts with the identification of probable precursors in the MS spectrum using accurately determined masses and proceeds with | ||
| − | identifying phosphorylcholine headgroup fragment in the MS/MS spectra | ||
| − | A query for a | + | A query for a phosphatidylcholine lipid (PC) could be: |
| − | |||
| − | |||
| − | |||
| − | + | *Find all precursor masses, which fit into the following set of sum compositions: "C[30..48] H[30..200] O[8] P[1] N[1]" and | |
| + | *look if there is the "C5 H15 O4 P1 N1" fragment (or m/z 184.07) in its MS/MS spectrum. | ||
| + | *if those two conditions hold, we identified a phosphatidylcholine and can report the lipid species | ||
| − | + | === MFQL identification of phosphatidylcholines (PC) === | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | [[Image:Figure6.png|center|600px|MFQL identification of phosphatidylcholines (PC)]] '''Figure:''' '''Identification of a PC lipid.''' Upon their collisional fragmentation, molecular cations of PC produce a specific head group fragment with m/z 184.07 and sum composition 'C5 H15 O4 P1 N1'. '''A:''' MS spectrum acquired by direct infusion of a total lipid extract into a QSTAR mass spectrometer (inset). All detectable peaks were subjected to MS/MS. The spectrum acquired from the precursor m/z 788.5 (designated by the arrow) is presented at the lower panel. The precursor ion was isolated within 1 Da mass range and therefore several isobaric lipid precursors were co-isolated for MS/MS and produced abundant fragment ions unrelated to PC. These ions were disregarded by this MFQL query and did not affect PC identification. '''B:''' MFQL query identifying PC species, details are provided in the text. '''C:''' screenshot of the output spreadsheet file; column annotation and content is determined by REPORT section of the above MFQL, see also text for details. | ||
| − | First, let us assign a name to the query: | + | <br> For better illustration of the structure of MFQL and the meaning of the different command lines we explain in the following the example script for identification of PC lipid specie. First, let us assign a name to the query: |
| − | <pre>QUERYNAME = Phosphatidylcholine;</pre> | + | <pre>QUERYNAME = Phosphatidylcholine;</pre> |
| − | Next, we define the variables used for identifying the species. | + | Next, we define the variables used for identifying the species. Our query should identify the singly charged PC head group fragment and therefore: |
| − | Our query should identify the singly charged PC head group | + | <pre>DEFINE |
| − | fragment and therefore: | ||
| − | <pre> | ||
| − | DEFINE | ||
headPC = 'C5 H15 O4 N1 P1' WITH CHG = +1; | headPC = 'C5 H15 O4 N1 P1' WITH CHG = +1; | ||
| − | </pre> | + | </pre> |
| − | The keyword <tt>CHG</tt> states the charge of the ion. | + | The keyword <tt>CHG</tt> states the charge of the ion. |
| − | In a shotgun experiment not all fragmented peaks will originate from PCs. | + | In a shotgun experiment not all fragmented peaks will originate from PCs. For higher search specificity we next define precursors (<tt>prPC</tt>), who are expected to produce <tt>headPC</tt> fragment in MS/MS spectra. We impose the sc-constraint on precursor masses: besides sum composition requirements, it requests that precursors are singly charged and their unsaturation (expressed as a double bond equivalent with the keyword <tt>DBR</tt>) is within a certain (here from 1.5 to 7.5) range: |
| − | For higher search specificity we next define precursors (<tt>prPC</tt>), who are expected | + | <pre>DEFINE |
| − | to produce <tt>headPC</tt> fragment in MS/MS spectra. We impose the sc-constraint on precursor | ||
| − | masses: besides sum composition requirements, it requests that precursors are singly | ||
| − | charged and their unsaturation (expressed as a double bond equivalent with the keyword | ||
| − | <tt>DBR</tt>) is within a certain (here from 1.5 to 7.5) range: | ||
| − | <pre> | ||
| − | DEFINE | ||
prPC = 'C[30..48] H[30..200] N[1] O[8] P[1]' WITH CHG = +1, DBR = (1.5, 7.5); | prPC = 'C[30..48] H[30..200] N[1] O[8] P[1]' WITH CHG = +1, DBR = (1.5, 7.5); | ||
| − | </pre> | + | </pre> |
| − | + | Next, the IDENTIFY section specifies that <tt>prPC</tt> precursors should be identified in MS spectra and <tt>headPC</tt> fragments in MS/MS spectra, both acquired in positive mode. The logical operation AND requests that <tt>headPC</tt> should only be searched in MS/MS spectra of <tt>prPC</tt> | |
| − | Next, the IDENTIFY section specifies that <tt>prPC</tt> precursors should be | + | <pre>IDENTIFY |
| − | identified in MS spectra and <tt>headPC</tt> fragments in MS/MS spectra, both | ||
| − | acquired in positive mode. The logical operation AND requests that <tt>headPC</tt> | ||
| − | should only be searched in MS/MS spectra of <tt>prPC</tt> | ||
| − | <pre> | ||
| − | IDENTIFY | ||
prPC IN MS1+ AND | prPC IN MS1+ AND | ||
headPC IN MS2+ | headPC IN MS2+ | ||
| − | </pre> | + | </pre> |
| − | We further limit the search space by applying optional project-specific | + | We further limit the search space by applying optional project-specific compositional constraints formulated in the next SUCHTHAT section. For example, it is generally assumed that mammals do not produce fatty acids having an odd number of carbon atoms. Therefore, it is likely that if a recognized lipid comprises an odd-numbered fatty acid moiety this identification is false. |
| − | compositional constraints formulated in the next SUCHTHAT section. For example, | + | <pre>SUCHTHAT |
| − | it is generally assumed that mammals do not produce fatty acids having an odd | ||
| − | number of carbon atoms. Therefore, it is likely that if a recognized lipid | ||
| − | comprises an odd-numbered fatty acid moiety this identification is false. | ||
| − | <pre> | ||
| − | SUCHTHAT | ||
isEven(prPC.chemsc[C]); | isEven(prPC.chemsc[C]); | ||
| − | </pre> | + | </pre> |
| − | In this case the operator <tt>isEven</tt> requests that candidate PC | + | In this case the operator <tt>isEven</tt> requests that candidate PC precursors should contain an even number of carbon atoms. Since the head group of PC and the glycerol backbone contain 5 and 3 carbon atoms, respectively, this implies that a lipid could not comprise fatty acid moieties with odd and even number of carbon atoms at the same time. By executing the DEFINE, IDENTIFY and SUCHTHAT sections LipidXplorer will recognize spectra pertinent to PC species. The last section REPORT defines how these findings will be reported. This includes annotation of the recognized lipid species, reporting the abundances of characteristic ions for subsequent quantification and reporting all additional information pertinent to the analysis, such as masses, mass differences (errors) etc. LipidXplorer outputs the findings as a *.csv file in which identified species are in rows, while the columns content is user-defined. In this example we define 5 columns: <tt>NAME</tt> - to report the species name; along with four peak attributes such as: <tt>MASS</tt> - species mass-to-charge ratio; <tt>CHEMSC</tt> - chemical sum composition; <tt>ERROR</tt> - the mass measurement error (the difference of the theoretical to the measured mass); <tt>INTENS</tt> - intensities of the specified ions reported for each individual acquisition. |
| − | precursors should contain an even number of carbon atoms. Since the head | + | <pre>REPORT |
| − | group of PC and the glycerol backbone contain 5 and 3 carbon atoms, | ||
| − | respectively, this implies that a lipid could not comprise fatty acid | ||
| − | moieties with odd and even number of carbon atoms at the same time. | ||
| − | By executing the DEFINE, IDENTIFY and SUCHTHAT sections LipidXplorer will | ||
| − | recognize spectra pertinent to PC species. The last section REPORT | ||
| − | defines how these findings will be reported. This includes annotation | ||
| − | of the recognized lipid species, reporting the abundances of characteristic | ||
| − | ions for subsequent quantification and reporting all additional | ||
| − | information pertinent to the analysis, such as masses, mass differences | ||
| − | (errors) etc. LipidXplorer outputs the findings as a *.csv file in which | ||
| − | identified species are in rows, while the columns content is user-defined. | ||
| − | In this example we define 5 columns: <tt>NAME</tt> - to report the species name; | ||
| − | along with four peak attributes such as: <tt>MASS</tt> - species mass; | ||
| − | <tt>CHEMSC</tt> - chemical sum composition; <tt>ERROR</tt> - difference | ||
| − | to the | ||
| − | ions reported for each individual acquisition. | ||
| − | <pre> | ||
| − | REPORT | ||
MASS = prPC.mass; | MASS = prPC.mass; | ||
| − | NAME = "PC [%d:%d]" % "((prPC.chemsc - headPC.chemsc)[C] - 3, prPC.chemsc[db] - 1.5)"; | + | NAME = "PC [%d:%d]" % "((prPC.chemsc - headPC.chemsc)[C] - 3, prPC.chemsc[db] - 1.5)"; |
CHEMSC = prPC.chemsc; | CHEMSC = prPC.chemsc; | ||
| − | ERROR = "% | + | ERROR = "%.2fppm" % "(prPC.errppm)"; |
INTENS = prPC.intensity; | INTENS = prPC.intensity; | ||
FRAGINTENS = headPC.intensity;; | FRAGINTENS = headPC.intensity;; | ||
| − | </pre> | + | </pre> |
| + | <br> It is also possible to define mathematical terms or use certain functions, such as text formatting, on these attributes. The text format implies two strings separated by <tt>%</tt> , where the first string contains placeholders and the second string their content. This formatting is used in the NAME string such that the actual annotation convention remains in the users discretion. In this example two placeholders <tt>%d</tt> of the lipids class name <tt>PC [%d:%d]</tt> are filled with the number of carbon atoms and double bonds in the fatty acid moieties. The number of the<span style="font-weight: bold;"> </span>carbon atoms is calculated by subtracting the <tt>headPC</tt> carbon atoms and the 3 carbons of the glycerol backbone from the total carbon of the precursor <tt>prPC</tt> (Figures 5 and 6). | ||
| + | == General rules in MFQL queries == | ||
| − | + | #Everything written after <tt>#</tt> is ignored by the interpreter. This function is used for writing comments in the code. | |
| − | + | #Every line has to end with <tt>;</tt> | |
| − | + | #Every query has to end with an extra <tt>;</tt> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | <br> | |
| − | + | == The structure of an MFQL query == | |
| − | |||
| − | |||
| + | A MFQL query consists of 3-4 sections: | ||
| − | + | 1. '''DEFINE''': defines sum compositions, sc-constraints (see also [[#sc-constraints]]), masses or groups of masses and associates them to user defined names.<br> | |
| − | |||
| − | + | 2. '''IDENTIFY''': determines where and how the DEFINE content is applied. It usually encompasses searches for specific precursors in MS and/or fragment ions and/or neutral losses in MS/MS spectra<br> | |
| − | |||
| − | + | 3. '''SUCHTHAT''': ''is optional''. It defines constraints that are formulated as mathematical expressions and inequalities, numerical values, peak attributes (see Supporting Information S-4), sum compositions and functions. Several individual constraints can be bundled by logical operations and applied together.<br> | |
| − | It | ||
| − | + | 4. '''REPORT''': establishes the content and format of the output <br> | |
| − | |||
| − | |||
| − | |||
| − | + | After '''REPORT''' there is a list of variables (<tt>MASS</tt>, <tt>NAME</tt>, ...) which represent columns in the output file. Each columns content is defined after the <tt>=</tt>. More on the '''REPORT''' will be found in the '''REPORT''' chapter. | |
| − | + | == SC-constrains == | |
| − | |||
| − | |||
| − | == | + | For dealing with sets of chemical formulas LipidXplorer uses a special format which is called sum composition constraint (sc-constraint). With sc-constraints it is possible to specify sets of chemical formulas of a lipid class. Here is an example: |
| + | <pre>'C[38..54] H[30..130] O[10] N[1] P[1]' WITH DBR=(2.5,9.5), CHG = -1;</pre> | ||
| + | *<tt><span style="font-family: sans-serif;">'C[38..54] .... P[1]' is the sc-constraint defining a set of chemical formulas</span></tt> | ||
| + | *<tt>DBR</tt> means 'Double Bond Range' and narrows the number of possible double bonds and rings to the given numbers. | ||
| + | *<tt>CHG</tt> states the charge. If the charge is set to zero then the sc-constraint will be threat as a collection of neutral losses. | ||
| − | + | == The 4 sections of a MFQL query == | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | === Part 1: Definition of sum composition, sc-constrains and masses === | |
| − | + | The first statement of any query is | |
| − | + | <pre>QUERYNAME = <name of the query></pre> | |
| + | to give the query a unique name. | ||
| − | == | + | Next, variables are defined. It's syntax is |
| + | <pre>DEFINE <variable name> = (<chemical sum composition> | <sc-constraint> | <mass>) (WITH (<option> = <value>)+)? | ||
| + | </pre> | ||
| + | After the keyword <tt>DEFINE</tt> comes the name of the variable followed by equation sign and its content. This can be either a chemical sum composition, a sc-constrain or a list of sum compositions. Sum compositions and sc-constraints are written in single quotes. Then there can be a <tt>WITH</tt> followed by certain options. The options can be: | ||
| − | + | #<tt>DBR</tt> is the double bound range of a sc-constrain. It is a 2-tuple stating the minimum and the maximum double bounds and rings which are allowed for a sum composition of this sc-constrain. | |
| + | #<tt>CHG</tt> states the charge | ||
| − | + | If the fragment should be a neutral loss, this can be stated by setting the charge to zero with <tt>CHG = 0</tt> or by writing <tt>AS NEUTRALLOSS</tt> after the sum composition or sc-constrain. | |
| − | < | ||
| − | |||
| − | + | NOTE: The neutral loss is calculated always between the precursor mass and the fragment, never between two fragments. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ==== examples ==== | |
| − | |||
| − | + | Define PC-O sc-constrains and PC-O's head group which is connected to the precursor mass: | |
| − | + | <pre>DEFINE PR = 'C[30..48] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | Define PC-O sc-constrains and PC-O's head group which is connected to the | ||
| − | precursor mass: | ||
| − | <pre> | ||
| − | DEFINE PR = 'C[30..48] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; | ||
DEFINE pcHead = 'C5 H15 O4 P1 N1' WITH CHG = 1; | DEFINE pcHead = 'C5 H15 O4 P1 N1' WITH CHG = 1; | ||
| − | </pre> | + | </pre> |
| − | + | Define PE sc-constrains and PE's head group which is connected to the precursor mass: | |
| − | Define PE sc-constrains and PE's head group which is connected to the | + | <pre>DEFINE PR = 'C[30..46] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; |
| − | precursor mass: | ||
| − | <pre> | ||
| − | DEFINE PR = 'C[30..46] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; | ||
DEFINE peHead = 'C2 H8 O4 N1 P1' AS NEUTRALLOSS; | DEFINE peHead = 'C2 H8 O4 N1 P1' AS NEUTRALLOSS; | ||
| − | </pre> | + | </pre> |
| − | + | Define sc-constrains and fragments for PE-Plasmalogen: | |
| − | Define sc-constrains and fragments for PE-Plasmalogen: | + | <pre>DEFINE PR = 'C[30..46] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; |
| − | <pre> | ||
| − | DEFINE PR = 'C[30..46] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; | ||
DEFINE FRAG1 = 'C[14..26] H[20..80] O[3]' WITH DBR = (1.5,9), CHG = 1; | DEFINE FRAG1 = 'C[14..26] H[20..80] O[3]' WITH DBR = (1.5,9), CHG = 1; | ||
DEFINE FRAG2 = 'C[14..26] H[20..80] N[1] O[4] P[1]' WITH DBR = (1.5,9), CHG = 1; | DEFINE FRAG2 = 'C[14..26] H[20..80] N[1] O[4] P[1]' WITH DBR = (1.5,9), CHG = 1; | ||
</pre> | </pre> | ||
| + | An arbitrary number of variables can be defined, but they are only valid for the current query. I.e. they are not valid in other queries of the same Run. | ||
| − | + | === Part 2: The <tt>IDENTIFY</tt> section === | |
| − | |||
| − | |||
| − | ===Part 2: The <tt>IDENTIFY</tt> section=== | ||
| − | The before defined variables are queried to the experiment database. The syntax is: | + | The before defined variables are queried to the experiment database. The syntax is: |
<pre>IDENTIFY | <pre>IDENTIFY | ||
| Line 250: | Line 131: | ||
... | ... | ||
<identification n> | <identification n> | ||
| − | </pre> | + | </pre> |
| + | The headline 'IDENTIFY' is followed by identifications which are connected by 'AND'. The result of an identification can be a singleton or a set, i.e. for some variables more than one mass is identified. This holds especially for sc-constraints. This section is the first filtering step. The section returns ''True'' if the boolean expression is true. The expression is true if the particular expressions are true: | ||
| − | + | An identification looks like this: | |
| − | + | <pre>((<variable name> IN (MS1+/-|MS2+/-)+)? | |
| − | An identification looks like this: | ||
| − | <pre> | ||
| − | ((<variable name> IN (MS1+/-|MS2+/- | ||
</pre> | </pre> | ||
| + | Here does LipidXplorer check the existence of certain masses/fragment masses. The scope (level of MS) is stated after 'IN': The 'MS1+', 'MS1-', 'MS2+' and 'MS2-' tags point to the MS level where to look for the sum composition ('MS1+' means in positive MS, while 'MS2-' means in negative MS/MS). | ||
| − | + | == Emulating (Multiple) Precursor Ion Scan / Neutral Loss Scan with MFQL == | |
| − | |||
| − | + | In the <tt>IDENTFIY</tt> section specify precursor ion scans (PIS) and neutral loss scans (NLS)can be defined. If the variable is a sc-constrain it emulates multiple PIS/NLS. Switching from PIS to NLS is done in the definition part. When a variable gets charge zero (<tt>CHG = 0</tt>) or the keyword <tt>AS NEUTRALLOSS</tt> is given then it is stated as neutral loss. Otherwise it is stated as (fragment) mass. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | (Comment: The above feature should not be not mistaken with the LipidXplorer functionality to import PIS and NLS mass spectrometric acquisitions.) | |
| − | |||
| − | |||
| − | |||
| − | |||
| + | Some examples: | ||
<pre># Phosphatedylcholine ether species | <pre># Phosphatedylcholine ether species | ||
DEFINE PR = 'C[30..48] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; | DEFINE PR = 'C[30..48] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; | ||
DEFINE pcHead = 'C5 H15 O4 P1 N1' WITH CHG = 1; | DEFINE pcHead = 'C5 H15 O4 P1 N1' WITH CHG = 1; | ||
| − | IDENTIFY | + | IDENTIFY |
# the MS mass should fit to 'PR' and it should have a MS/MS fragment mass fitting to 'pcHead' | # the MS mass should fit to 'PR' and it should have a MS/MS fragment mass fitting to 'pcHead' | ||
| − | PR IN MS1+ | + | PR IN MS1+ AND |
| − | + | pcHead in MS2+ | |
| − | pcHead in MS2+ | ||
################################################################################ | ################################################################################ | ||
| Line 292: | Line 162: | ||
DEFINE peHead = 'C2 H8 O4 N1 P1' WITH CHG = 0; | DEFINE peHead = 'C2 H8 O4 N1 P1' WITH CHG = 0; | ||
| − | IDENTIFY | + | IDENTIFY |
# marking | # marking | ||
| − | PR IN MS1+ | + | PR IN MS1+ AND |
| − | peHead in MS2+ | + | peHead in MS2+ |
################################################################################ | ################################################################################ | ||
| Line 305: | Line 175: | ||
DEFINE FRAG2 = 'C[14..26] H[20..80] N[1] O[4] P[1]' WITH DBR = (1.5,9), CHG = 1; | DEFINE FRAG2 = 'C[14..26] H[20..80] N[1] O[4] P[1]' WITH DBR = (1.5,9), CHG = 1; | ||
| − | IDENTIFY | + | IDENTIFY |
# marking | # marking | ||
| − | PR IN MS1+ | + | PR IN MS1+ AND |
| − | FRAG1 IN MS2+ | + | FRAG1 IN MS2+ AND |
| − | FRAG2 IN MS2+ | + | FRAG2 IN MS2+ |
| − | </pre> | + | </pre> |
| + | === Part 3: The <tt>SUCHTHAT</tt> section === | ||
| − | + | After the collection of specific masses, it is possible to add more constraints to the query. For example: the identification of PE Plasmalogen requires the marking of 'FRAG1' and 'FRAG2' which both contain several possibilities since they are sc-constraints (see example above) and a test if those two fragments in sum match the precursor mass, i.e. is "FRAG1 + FRAG2 == PR"? Such a constraint is formulated in the optional 'SUCHTHAT' section as boolean connected equations, unequations and functions. The syntax is: | |
| − | |||
| − | After the collection of specific masses, it is possible to add more constraints to the query. For example: the identification of PE Plasmalogen requires the marking of 'FRAG1' and 'FRAG2' which both contain several possibilities since they are sc-constraints (see example above) and a test if those two fragments in sum match the precursor mass, i.e. is "FRAG1 + FRAG2 == PR"? Such a constraint is formulated in the optional 'SUCHTHAT' section as boolean connected equations, unequations and functions. The syntax is: | ||
<pre>SUCHTHAT | <pre>SUCHTHAT | ||
(((NOT)? (<equation> | <unequation> | <function>)) | | (((NOT)? (<equation> | <unequation> | <function>)) | | ||
((NOT)? (<equation> | <unequation> | <function>) (AND | OR))+) (WITH (<option> = <value>)+)? | ((NOT)? (<equation> | <unequation> | <function>) (AND | OR))+) (WITH (<option> = <value>)+)? | ||
</pre> | </pre> | ||
| − | The terms can be build up with the basic mathematical functions +, -, *, /. Parenthesis can also be used. The terms are connected as equations by '==' and as inequalities by ' | + | The terms can be build up with the basic mathematical functions +, -, *, /. Parenthesis can also be used. The terms are connected as equations by '==' and as inequalities by '<', '>', '<=', '>=' and '!=' for not equal. The values for the terms can be marked masses (given with their variable name), floating point numbers or chemical sum compositions. Certain attributes of marked masses can be also addressed. This can be done by writing the attribute after the variable name connected with a dot. The intensity of the peak 'PR' for example is addressed as <tt>PR.intensity</tt>. A list of peak attributes can be found here: [[#List_of_peak_attributes]] |
| − | The values for the terms can be marked masses (given with their variable name), floating point numbers or chemical sum compositions. Certain attributes of marked masses can be also addressed. This can be done by writing the attribute after the variable name connected with a dot. The intensity of the peak 'PR' for example is addressed as <tt>PR.intensity</tt>. A list of peak attributes can be found here: [[# | ||
| − | ====Functions==== | + | ==== Functions ==== |
| − | Additional to the attributes, SUCHTHAT supports the use of functions. The list of all functions can be found here: [[# | + | Additional to the attributes, SUCHTHAT supports the use of functions. The list of all functions can be found here: [[#List_of_functions]] |
| − | ===Part 4: The <tt>REPORT</tt> section=== | + | === Part 4: The <tt>REPORT</tt> section === |
| − | All successful identifications are piped to the <tt>REPORT</tt> section, | + | All successful identifications are piped to the <tt>REPORT</tt> section, where the format of the output is specified. In general the <tt>REPORT</tt> consists of a list of variables where each represents a column. The content of the variable is the content of the column. So is the following code generates a column with the name <tt>MASS</tt> and the m/z values of <tt>PR</tt>'s identified species as content: |
| − | where the format of the output is specified. In general the <tt>REPORT</tt> | + | <pre>REPORT |
| − | consists of a list of variables where each represents a column. The content | ||
| − | of the variable is the content of the column. So is the following code | ||
| − | generates a column with the name <tt>MASS</tt> and the m/z values of <tt>PR</tt>'s | ||
| − | identified species as content: | ||
| − | <pre> | ||
| − | REPORT | ||
MASS = PR.mass | MASS = PR.mass | ||
| − | </pre> | + | </pre> |
| − | + | The next example reports the sum of the intensities of two fragments | |
| − | The next example reports the sum of the intensities of two fragments | + | <pre>REPORT |
| − | <pre> | ||
| − | REPORT | ||
INTENS = frag1.intensity + frag2.intensity | INTENS = frag1.intensity + frag2.intensity | ||
| − | </pre> | + | </pre> |
| − | + | Mostly those fragments can be the same (so for example for 2 fatty acid scans), therefore LipidXplorer has a special function which does not sum intensities of same fragments: | |
| − | Mostly those fragments can be the same (so for example for 2 fatty acid scans), therefore LipidXplorer has a special function which does not sum intensities of same fragments: | + | <pre>REPORT |
| − | <pre> | + | INTENS = sumIntensity(frag1.intensity, frag2.intensity) |
| − | REPORT | + | </pre> |
| − | INTENS = sumIntensity(frag1, frag2) | + | The syntax of <tt>REPORT</tt> is: |
| − | </pre> | ||
| − | |||
| − | The syntax of <tt>REPORT</tt> is: | ||
<pre>REPORT | <pre>REPORT | ||
((<variable name> = <variable> | <equation>) | ((<variable name> = <variable> | <equation>) | ||
| − | </pre> | + | </pre> |
| + | The content of the variable can be any attribute and/or term as in the <tt>SUCHTHAT</tt> section. The <tt>REPORT</tt> section has an additional feature with which it is possible to generate lipid names or other formatted strings. | ||
| − | The | + | The syntax for this function is: |
| − | <tt> | + | <pre>REPORT |
| − | + | (<variable name> = "<format string>" % ((<list of variables for the format string>)+) | |
| − | + | </pre> | |
| − | + | The string format works as follows: there are two strings to give which are separated with a <tt>%</tt>. The first string contains the output format, i.e. a string with placeholders. Placeholder can be: <tt>%d</tt> for decimal values, <tt>%.</tt>''n''<tt>f</tt> for floating point values with ''n'' decimals and <tt>%s</tt> for string values. The second string contains a list with the content of the placeholders according to their order. For example: | |
<pre>REPORT | <pre>REPORT | ||
| − | + | LIPIDNAME = "PC [%d:%d]" % (fa1PC.chemsc[C] + fa2PC.chemsc[C], fa1PC.chemsc[db] + fa2PC.chemsc[db]) | |
</pre> | </pre> | ||
| + | The variable <tt>LIPIDNAME</tt> contains the string <tt>"PC [... : ...]"</tt>. The first decimal value is filled with the sum of the carbon atoms of both fatty acids <tt>(fa1PC, fa2PC)</tt> and the second decimal value the sum of the double bonds. The output could be for example <tt>"PC [36:2]"</tt>. | ||
| − | The string | + | The format string variant is a Python gimmick, where MFQL uses standard Python commands. I.e. the format string is a python function (see [http://docs.python.org/library/stdtypes.html#string-formatting-operations here] for more information). |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | === Notes === | |
| − | |||
| − | |||
| − | + | *If a lipid was not found in a particular sample, its intensity is set to zero. | |
| + | *If the isotopic correction corrects an intensity to zero or less than zero, it is set to '-1' | ||
| − | + | == List of peak attributes == | |
| − | |||
| − | == | + | ==== error ==== |
| − | |||
The difference between the theoretical mass (according to the sum composition) and the tagged mass from the spectrum. The error can be given in the 3 types: | The difference between the theoretical mass (according to the sum composition) and the tagged mass from the spectrum. The error can be given in the 3 types: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | #<tt>errppm</tt> -> error in ppm |
| + | #<tt>errda</tt> -> error in dalton | ||
| + | #<tt>errres</tt> -> error as resolution value | ||
| + | |||
| + | ==== mass ==== | ||
| + | |||
| + | The m/z value of the peak | ||
| + | |||
| + | ==== chemsc ==== | ||
| + | |||
| + | The chemical sum composition. For addressing certain elements of the sum composition, the element is to write in brackets after <tt>.chemsc</tt>. To get the number of <tt>C</tt> atoms from a formula for example: | ||
| + | <pre>PR.chemsc[C]</pre> | ||
| + | #<tt>frsc</tt> -> the chemical sum composition of the fragment. If the peak is defined as a (charged) fragment, it is the same as <tt>chemsc</tt>, if it is defined as a neutral loss, it returns the sum composition of the fragment. | ||
| + | #<tt>nlsc</tt> -> the chemical sum composition of the neutral loss. If the peak is defined as (uncharged) a neutral loss, it is the same as <tt>chemsc</tt>, if it is defined as a fragment, it returns the sum composition of the neutral loss of the precursor. | ||
| + | |||
| + | ==== intensity ==== | ||
| + | |||
| + | All the intensities of a mass from all the samples it occured. Note that <tt>intensity</tt> is mostly no single value but a list of intensities. One list entry for every sample the peak was found. If used in an equation or unequation, the whole list is considered. I.e. PR.intensity > 10000 is true if and only if all intensities are greater than 10000. It is possible to address only a part of all samples. This is done by writing the name of the sample group as string with wildcards (<tt>*</tt> and/or <tt>?</tt>). E.g. is <tt>PR.intensity["*blanck*"]</tt> returning just the samples with the string <tt>blanck</tt> in their name. This could be all blanck samples. This feature allows to generate sample groups by naming the samples according to their group. So, a lot of different constraints can be stated, which increase the accuracy of the interpretation or even already interpret the result. E.g. | ||
| + | <pre> avg(PR.intensity["*blanck*"]) < avg(PR.intensity["*exp*"]) / 100 </pre> | ||
| + | This statement asserts that the one percent of the average intensity of all experimental samples ("*exp*") should be greater than the average intensity found in the blanck sample. This simply throws out every "lipid", which is obviously noise. | ||
| + | |||
| + | ==== binsize ==== | ||
| + | |||
| + | The size of the bin of the peak coming from the averaging algorithm. The value is given in Dalton. | ||
| + | |||
| + | ==== occ ==== | ||
| − | = | + | Is the occupation of the peak. Occupation = nb. of occurences in the sample / nb. of samples |
| − | + | == List of functions == | |
| − | ==== | + | ==== isEven(n) ==== |
| − | where n is an integer value. The function returns True, if n is | + | where n is an integer value. The function returns True, if n is even. E.g.: <tt>isEven(PR.chemsc[C])</tt>. |
| − | ==== | + | ==== isOdd(n) ==== |
| − | where n is | + | where n is an integer value. The function returns True, if n is odd. |
| − | ==== | + | ==== avg(v.intensity) ==== |
| − | where | + | where n is a variable. The function returns the average value of the intensities of n. E.g.: |
| + | <pre>avg(PR.intensity)</pre> | ||
| + | ==== isStandard(v, scope) ==== | ||
| − | + | where v is a variable and scope is "MS1+", "MS1-", "MS2+" or "MS2-". This function is special since it does not return anything. It enables the automatic calculation of standardizied intensities according to the given standard in v. I.e. Every intensity is calculated as relative to v. | |
| − | + | ==== sumIntensity(f1.intensity, f2.intensity, ...) ==== | |
| − | |||
| − | |||
| − | |||
| − | + | The function sumIntensity() is used for summing up intensities of different MS2 entries where multiple peaks are required for identification and quantification. In case of fragments with isotopic corrected place holders (see above)the following rules were implemented. | |
| − | + | If all MasterScan entries in the MS2 for a particular molecule are place holders (i.e. all are set to '-1') then those values are just added and will result in <math>n_i\times -1</math> where <span class="texhtml">''n''<sub>''i''</sub></span> is the number of the attributes. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | If there is just one entry whose intensity is greater zero all <span class="texhtml"> − 1</span> place holders are threaded as zero and not added to the overall sum. In the presented example we assume that two entries in the MS2 where used for the sumIntensity() function: | |
| − | + | <span class="texhtml">''F''1 + ''F''2 − > sumIntensity(''F''1.intensity,''F''2.intensity)</span> | |
| + | <span class="texhtml"> − 1 + − 1 = − 2</span> | ||
| + | <span class="texhtml">0 + − 1 = − 1</span> | ||
| + | <span class="texhtml">1 + − 1 = 1</span> | ||
| + | <span class="texhtml">2 + − 1 = 2</span> | ||
| + | <span class="texhtml">2 + 0 = 2</span> | ||
| − | + | That has following consequences when such results have to be interpreted: | |
| − | + | A) intensity = 0 in this specific sample none of the required fragments was present | |
| − | + | B) intensity < 0 in this sample some of the required fragments were found in the initial MasterScan but set '-1', none fragment above threshold (1) was present | |
| − | == | + | C) intensity = -<span class="texhtml">''n''<sub>''i''</sub></span> all fragments were below the threshold (1) after isotopic correction |
| + | D) intensity > 0 in this case at least one of the required fragments was after isotopic correction above the threshold (1) | ||
| + | |||
| + | === Some examples === | ||
<pre>SUCHTHAT | <pre>SUCHTHAT | ||
# the number of 'C' atoms in 'PR's chemical sum composition should be odd | # the number of 'C' atoms in 'PR's chemical sum composition should be odd | ||
| Line 465: | Line 318: | ||
SUCHTHAT | SUCHTHAT | ||
# the sum of both fragments ('FRAG1', 'FRAG2') minus one 'H' should be equal to | # the sum of both fragments ('FRAG1', 'FRAG2') minus one 'H' should be equal to | ||
| − | # the precursor mass ('PR') | + | # the precursor mass ('PR') and |
# the intensity of 'FRAG2' should be bigger than 3/10th of the | # the intensity of 'FRAG2' should be bigger than 3/10th of the | ||
# the intensity of 'FRAG1' | # the intensity of 'FRAG1' | ||
| − | FRAG1 + FRAG2 - 'H1' == PR | + | FRAG1 + FRAG2 - 'H1' == PR AND |
FRAG1.intensity * 3 < FRAG2.intensity * 10 | FRAG1.intensity * 3 < FRAG2.intensity * 10 | ||
</pre> | </pre> | ||
| − | == The principle of the | + | == How LipidXplorer runs multiple MFQL queries == |
| + | |||
| + | The principle of a LipidXplorer Run is the following: All queries run successively on the given MasterScan. For every query, LipidXplorer iterates through the list of MS masses of the MasterScan from smallest to the greatest and checks the conditions given in definition, <tt>IDENTIFY</tt>, <tt>SUCHTHAT</tt> and <tt>REPORT</tt> sections. I.e. | ||
| + | |||
| + | *it loads a MS mass | ||
| + | *it checks if it fits a given sum compostion or sc-constrain (definition and <tt>IDENTIFY</tt> section). | ||
| + | *it looks into its MS/MS spectrum (if provided) and does the same (definition and <tt>IDENTIFY</tt> section). | ||
| + | *the boolean constraints are checked (<tt>SUCHTHAT</tt> section) and if the result is | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
positive the MS mass is accepted and send to the <tt>REPORT</tt> section | positive the MS mass is accepted and send to the <tt>REPORT</tt> section | ||
| − | + | <br> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | ==Examples== | + | == Examples == |
| − | ===Screen (without MS/MS experiments) for Phosphatidylcholine species=== | + | === Screen (without MS/MS experiments) for Phosphatidylcholine species === |
| − | A "screen" is a fast identification based on only MS information. To do | + | A "screen" is a fast identification based on only MS information. To do screening properly the masses should be high accurate, because otherwise the error of identification is too high. |
| − | screening properly the masses should be high accurate, because otherwise | ||
| − | the error of identification is too high. | ||
| − | The name of the query here is <tt>Phosphatidylcholine</tt>. Giving a name | + | The name of the query here is <tt>Phosphatidylcholine</tt>. Giving a name to a query is obligatory and has to be done for every query. We define the sc-constraint <tt>prPC</tt> (short for "precursor of PC") and state that it should be found in the positive MS spectra. |
| − | to a query is obligatory and has to be done for every query. We define | ||
| − | the sc-constraint <tt>prPC</tt> (short for "precursor of PC") and state | ||
| − | that it should be found in the positive MS spectra. | ||
| − | Names for variables are arbitrary. The user should try to give meaningful | + | Names for variables are arbitrary. The user should try to give meaningful names in order to understand his query better. |
| − | names in order to understand his query better. | ||
| − | The <tt>IDENTIFIY</tt> section urges LipidXplorer to look for the precursor mass | + | The <tt>IDENTIFIY</tt> section urges LipidXplorer to look for the precursor mass into the MS spectrum. |
| − | into the MS spectrum. | ||
| − | In <tt>SUCHTHAT</tt> we use a function to restrict the result to lipids | + | In <tt>SUCHTHAT</tt> we use a function to restrict the result to lipids having an overall even number of carbon atoms. This means that the fatty acids of the lipid have to have both fatty acids even numbered or both odd numbered. Such, we can sort out lipids which we know they should not be in the organism we examine. |
| − | having an overall even number of carbon atoms. This means that the fatty | ||
| − | acids of the lipid have to have both fatty acids even numbered or | ||
| − | both odd numbered. Such, we can sort out lipids which we know they should | ||
| − | not be in the organism we examine. | ||
| − | The <tt>REPORT</tt> section uses the following variables: | + | The <tt>REPORT</tt> section uses the following variables: |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | *'MASS' returns the m/z value of the MS mass | ||
| + | *'NAME' returns the lipid species' name, which consists of the number of carbon atoms and double bonds of the fatty acids. Those numbers we get from taking the number of carbons/double bonds from the sum composition (prPC.chemsc[C]/prPC.chemsc[db]) and reduce it by the carbons/double bonds belonging to the PC's head group and glycerol backbone. | ||
| + | *'CHEMSC' returns the chemical sum composition | ||
| + | *'INTENS' returns the abundance of the identified lipid species for all samples | ||
| + | *'ERROR' returns the error of the finding in ppm. | ||
<pre>########################################################## | <pre>########################################################## | ||
# Identify PC with checking the precursor mass # | # Identify PC with checking the precursor mass # | ||
| Line 541: | Line 376: | ||
REPORT | REPORT | ||
MASS = prPC.mass; | MASS = prPC.mass; | ||
| − | NAME = "PC [%d:%d]" % | + | NAME = "PC [%d:%d]" % (prPC.chemsc[C] - 8, prPC.chemsc[db] - 5); |
CHEMSC = prPC.chemsc; | CHEMSC = prPC.chemsc; | ||
INTENS = prPC.intensity; | INTENS = prPC.intensity; | ||
| − | ERROR = "%2.2fppm" % | + | ERROR = "%2.2fppm" % (prPC.errppm); ; |
################ end script ################## | ################ end script ################## | ||
| − | </pre> | + | </pre> |
| + | The output of the query is the following: | ||
| − | + | [[Image:Screenshot-output.png|center|600px|OuputScreenShot]] | |
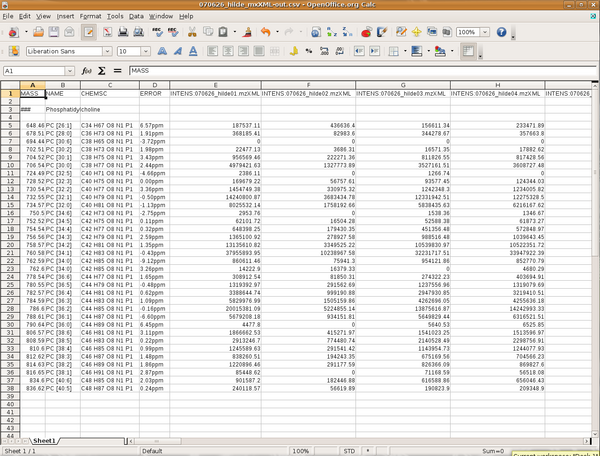
| − | + | This is a screen shot of spread sheet software holding the resulting data from the query. At the top are the variable names followed by the name of the query, then comes the content. Note, that for 'INTENS' the file name from which the sample data was taken is also written. Every entry in the result fulfills the constraints given in the query. If an expected value is not found then the query or the import settings should be refined. | |
| − | + | === Analysis of Phosphatidylcholine lipid species emulating PIS 184 === | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | === | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | Additionally to the former query we have a variable 'headPC' which contains the sum composition of the specific head group for PC which is found in the fragment spectra after MS/MS of a PC species. This variable is added as constraint in <tt>IDENTIFY</tt>. Thus a lipid is only identified if it fits to the constraints of <tt>prPC</tt> <tt>AND</tt> has a <tt>headPC</tt> fragment in its MS/MS spectrum. Again, we test the even numbers of carbons in <tt>SUCHTHAT</tt>, which ensure we do not find borderline masses, which actually cannot be in the sample. In the output we have additionally the abundance of the head group fragment with <tt>FRAGINTENS</tt>. | ||
<pre>########################################################## | <pre>########################################################## | ||
# Identify PCs with checking the precursor mass # | # Identify PCs with checking the precursor mass # | ||
| Line 596: | Line 413: | ||
REPORT | REPORT | ||
MASS = prPC.mass; | MASS = prPC.mass; | ||
| − | NAME = "PC [%d:%d]" % | + | NAME = "PC [%d:%d]" % ((prPC.chemsc - headPC.chemsc)[C] - 3, prPC.chemsc[db] - 1.5); |
CHEMSC = prPC.chemsc; | CHEMSC = prPC.chemsc; | ||
| − | ERROR = "%2.2fppm" % | + | ERROR = "%2.2fppm" % (prPC.errppm); |
INTENS = prPC.intensity; | INTENS = prPC.intensity; | ||
FRAGINTENS = headPC.intensity;; | FRAGINTENS = headPC.intensity;; | ||
| Line 605: | Line 422: | ||
</pre> | </pre> | ||
| − | === | + | === Application of Boolean operation "AND" for identification of PE-plasmalogen === |
| − | An example for a whole script: | + | An example for a whole script: |
<pre>########################################################### | <pre>########################################################### | ||
##### find PE-plasmalogens with MS2 in positive mode ###### | ##### find PE-plasmalogens with MS2 in positive mode ###### | ||
| Line 617: | Line 434: | ||
DEFINE FRAG2 = 'C[14..26] H[20..80] N[1] O[4] P[1]' WITH DBR = (1.5,9), CHG = 1; | DEFINE FRAG2 = 'C[14..26] H[20..80] N[1] O[4] P[1]' WITH DBR = (1.5,9), CHG = 1; | ||
| − | IDENTIFY | + | IDENTIFY |
# marking | # marking | ||
PR IN MS1+ AND | PR IN MS1+ AND | ||
| − | FRAG1 IN MS2+ | + | FRAG1 IN MS2+ AND |
| − | FRAG2 IN MS2+ | + | FRAG2 IN MS2+ |
SUCHTHAT | SUCHTHAT | ||
# the sum of both fragments ('FRAG1', 'FRAG2') minus one 'H' should be equal to | # the sum of both fragments ('FRAG1', 'FRAG2') minus one 'H' should be equal to | ||
| − | # the precurosor mass ('PR') | + | # the precurosor mass ('PR') and |
# the intensity of 'FRAG2' should be bigger than 3/10th of the | # the intensity of 'FRAG2' should be bigger than 3/10th of the | ||
# the intensity of 'FRAG1' | # the intensity of 'FRAG1' | ||
| − | FRAG1 + FRAG2 - 'H1' == PR | + | FRAG1 + FRAG2 - 'H1' == PR AND |
FRAG1.intensity * 3 < FRAG2.intensity * 10 | FRAG1.intensity * 3 < FRAG2.intensity * 10 | ||
| Line 639: | Line 456: | ||
# second is the lipids name generated with Python's string formatting function | # second is the lipids name generated with Python's string formatting function | ||
| − | NAME = "PE-O [%d:%dp / %d:%d]" % | + | NAME = "PE-O [%d:%dp / %d:%d]" % (FRAG1.frsc[C], FRAG1.frsc[db] - 2, FRAG2.frsc[C], FRAG2.frsc[db] - 2), |
# third is the precursor's chemical sum composition | # third is the precursor's chemical sum composition | ||
| Line 651: | Line 468: | ||
</pre> | </pre> | ||
| − | ==More Examples== | + | == More Examples == |
| − | More examples can be found in the MFQL collection provided in | + | More examples can be found in the [https://wiki.mpi-cbg.de/wiki/lipidx/index.php/Articles_Using_LipidXplorer MFQL collection] provided in the [https://wiki.mpi-cbg.de/wiki/lipidx/index.php/Main_Page LipidXplorer wiki]. |
| − | the LipidXplorer wiki. | ||
Latest revision as of 10:52, 24 July 2013
Contents
- 1 Introduction
- 2 A short tutorial
- 3 General rules in MFQL queries
- 4 The structure of an MFQL query
- 5 SC-constrains
- 6 The 4 sections of a MFQL query
- 7 Emulating (Multiple) Precursor Ion Scan / Neutral Loss Scan with MFQL
- 8 List of peak attributes
- 9 List of functions
- 10 How LipidXplorer runs multiple MFQL queries
- 11 Examples
- 12 More Examples
Introduction
MFQL is the first query language developed for the identification of molecules in complex shotgun spectra datasets. It formalizes the available or assumed knowledge of lipid fragmentation pathways into queries that are used for probing a MasterScan database.
Structural complexity of lipid species and sum composition constraints
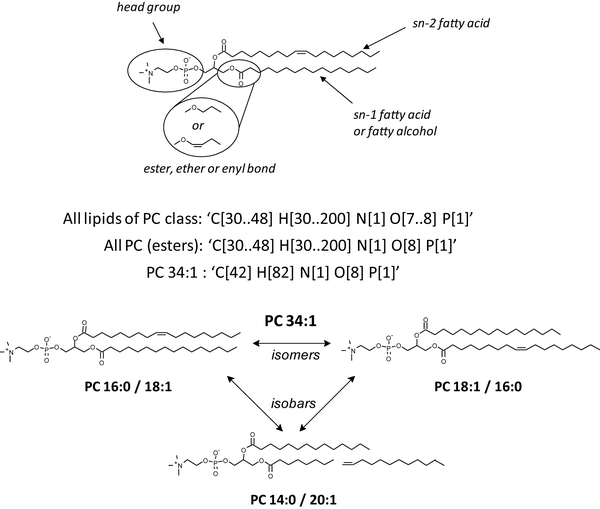
The figure shows the basic lipid structure and some characteristics specific for lipids using the example of a PC species. Let us consider PC as a representative example: PC molecules consist of a posphorylcholine head group attached to the glycerol backbone at the sn-3 position, while fatty acid moieties occupy sn-1 and sn-2 positions (alternatively, a fatty alcohol moiety could be attached at the sn-1 position). Fatty acid moieties differ by the number of carbon atoms and double bonds, but also by the relative location at the glycerol backbone, so that isomeric structures having exactly the same fatty acid moieties are possible. Note that isomeric structures are always isobaric, whereas isobaric molecules are not necessarily isomeric. Most generic constraints ("All lipids of PC class" or "All PC esters") encompass sum compositions of species with common naturally occurring fatty acids. However, because of the fatty acid variability, some species of other lipid classes (such as, PE) might meet the same constraint. Therefore, for most common glycerophospholipid classes, the characterization of individual molecular species could not solely rely on their intact masses, irrespective of how accurately were they measured. MS/MS experiments that produce structure-specific ions contribute more specific constraints, such as the number of carbons and double bonds in individual moieties, characteristic head group fragment, characteristic loss of a fatty acid moiety, among others. Within a MFQL query, these constraints can be bundled by Boolean operations.A short tutorial
Below we present an example of composing a MFQL query for identifying PC lipids in a typical shotgun dataset.
In MS/MS experiments (see #MFQL_identification_of_phosphatidylcholines_.28PC.29), molecular cations of PC species produce specific phosphorylcholine fragments of their head group having the sum composition of 'C5 H15 O4 N1 P1' and m/z 184.07 (see #MFQL_identification_of_phosphatidylcholines_.28PC.29). The identification of PC species starts with the identification of probable precursors in the MS spectrum using the accurately determined masses and proceeds with identifying phosphorylcholine headgroup fragment in the MS/MS spectra (see #MFQL_identification_of_phosphatidylcholines_.28PC.29).
A query for a phosphatidylcholine lipid (PC) could be:
- Find all precursor masses, which fit into the following set of sum compositions: "C[30..48] H[30..200] O[8] P[1] N[1]" and
- look if there is the "C5 H15 O4 P1 N1" fragment (or m/z 184.07) in its MS/MS spectrum.
- if those two conditions hold, we identified a phosphatidylcholine and can report the lipid species
MFQL identification of phosphatidylcholines (PC)
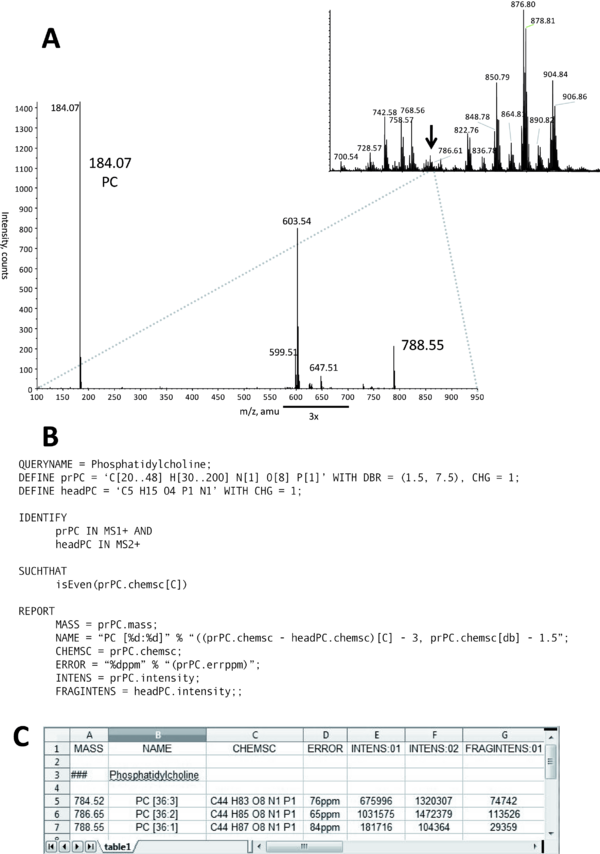
Figure: Identification of a PC lipid. Upon their collisional fragmentation, molecular cations of PC produce a specific head group fragment with m/z 184.07 and sum composition 'C5 H15 O4 P1 N1'. A: MS spectrum acquired by direct infusion of a total lipid extract into a QSTAR mass spectrometer (inset). All detectable peaks were subjected to MS/MS. The spectrum acquired from the precursor m/z 788.5 (designated by the arrow) is presented at the lower panel. The precursor ion was isolated within 1 Da mass range and therefore several isobaric lipid precursors were co-isolated for MS/MS and produced abundant fragment ions unrelated to PC. These ions were disregarded by this MFQL query and did not affect PC identification. B: MFQL query identifying PC species, details are provided in the text. C: screenshot of the output spreadsheet file; column annotation and content is determined by REPORT section of the above MFQL, see also text for details.
For better illustration of the structure of MFQL and the meaning of the different command lines we explain in the following the example script for identification of PC lipid specie. First, let us assign a name to the query:
QUERYNAME = Phosphatidylcholine;
Next, we define the variables used for identifying the species. Our query should identify the singly charged PC head group fragment and therefore:
DEFINE headPC = 'C5 H15 O4 N1 P1' WITH CHG = +1;
The keyword CHG states the charge of the ion.
In a shotgun experiment not all fragmented peaks will originate from PCs. For higher search specificity we next define precursors (prPC), who are expected to produce headPC fragment in MS/MS spectra. We impose the sc-constraint on precursor masses: besides sum composition requirements, it requests that precursors are singly charged and their unsaturation (expressed as a double bond equivalent with the keyword DBR) is within a certain (here from 1.5 to 7.5) range:
DEFINE prPC = 'C[30..48] H[30..200] N[1] O[8] P[1]' WITH CHG = +1, DBR = (1.5, 7.5);
Next, the IDENTIFY section specifies that prPC precursors should be identified in MS spectra and headPC fragments in MS/MS spectra, both acquired in positive mode. The logical operation AND requests that headPC should only be searched in MS/MS spectra of prPC
IDENTIFY prPC IN MS1+ AND headPC IN MS2+
We further limit the search space by applying optional project-specific compositional constraints formulated in the next SUCHTHAT section. For example, it is generally assumed that mammals do not produce fatty acids having an odd number of carbon atoms. Therefore, it is likely that if a recognized lipid comprises an odd-numbered fatty acid moiety this identification is false.
SUCHTHAT
isEven(prPC.chemsc[C]);
In this case the operator isEven requests that candidate PC precursors should contain an even number of carbon atoms. Since the head group of PC and the glycerol backbone contain 5 and 3 carbon atoms, respectively, this implies that a lipid could not comprise fatty acid moieties with odd and even number of carbon atoms at the same time. By executing the DEFINE, IDENTIFY and SUCHTHAT sections LipidXplorer will recognize spectra pertinent to PC species. The last section REPORT defines how these findings will be reported. This includes annotation of the recognized lipid species, reporting the abundances of characteristic ions for subsequent quantification and reporting all additional information pertinent to the analysis, such as masses, mass differences (errors) etc. LipidXplorer outputs the findings as a *.csv file in which identified species are in rows, while the columns content is user-defined. In this example we define 5 columns: NAME - to report the species name; along with four peak attributes such as: MASS - species mass-to-charge ratio; CHEMSC - chemical sum composition; ERROR - the mass measurement error (the difference of the theoretical to the measured mass); INTENS - intensities of the specified ions reported for each individual acquisition.
REPORT MASS = prPC.mass; NAME = "PC [%d:%d]" % "((prPC.chemsc - headPC.chemsc)[C] - 3, prPC.chemsc[db] - 1.5)"; CHEMSC = prPC.chemsc; ERROR = "%.2fppm" % "(prPC.errppm)"; INTENS = prPC.intensity; FRAGINTENS = headPC.intensity;;
It is also possible to define mathematical terms or use certain functions, such as text formatting, on these attributes. The text format implies two strings separated by % , where the first string contains placeholders and the second string their content. This formatting is used in the NAME string such that the actual annotation convention remains in the users discretion. In this example two placeholders %d of the lipids class name PC [%d:%d] are filled with the number of carbon atoms and double bonds in the fatty acid moieties. The number of the carbon atoms is calculated by subtracting the headPC carbon atoms and the 3 carbons of the glycerol backbone from the total carbon of the precursor prPC (Figures 5 and 6).
General rules in MFQL queries
- Everything written after # is ignored by the interpreter. This function is used for writing comments in the code.
- Every line has to end with ;
- Every query has to end with an extra ;
The structure of an MFQL query
A MFQL query consists of 3-4 sections:
1. DEFINE: defines sum compositions, sc-constraints (see also #sc-constraints), masses or groups of masses and associates them to user defined names.
2. IDENTIFY: determines where and how the DEFINE content is applied. It usually encompasses searches for specific precursors in MS and/or fragment ions and/or neutral losses in MS/MS spectra
3. SUCHTHAT: is optional. It defines constraints that are formulated as mathematical expressions and inequalities, numerical values, peak attributes (see Supporting Information S-4), sum compositions and functions. Several individual constraints can be bundled by logical operations and applied together.
4. REPORT: establishes the content and format of the output
After REPORT there is a list of variables (MASS, NAME, ...) which represent columns in the output file. Each columns content is defined after the =. More on the REPORT will be found in the REPORT chapter.
SC-constrains
For dealing with sets of chemical formulas LipidXplorer uses a special format which is called sum composition constraint (sc-constraint). With sc-constraints it is possible to specify sets of chemical formulas of a lipid class. Here is an example:
'C[38..54] H[30..130] O[10] N[1] P[1]' WITH DBR=(2.5,9.5), CHG = -1;
- 'C[38..54] .... P[1]' is the sc-constraint defining a set of chemical formulas
- DBR means 'Double Bond Range' and narrows the number of possible double bonds and rings to the given numbers.
- CHG states the charge. If the charge is set to zero then the sc-constraint will be threat as a collection of neutral losses.
The 4 sections of a MFQL query
Part 1: Definition of sum composition, sc-constrains and masses
The first statement of any query is
QUERYNAME = <name of the query>
to give the query a unique name.
Next, variables are defined. It's syntax is
DEFINE <variable name> = (<chemical sum composition> | <sc-constraint> | <mass>) (WITH (<option> = <value>)+)?
After the keyword DEFINE comes the name of the variable followed by equation sign and its content. This can be either a chemical sum composition, a sc-constrain or a list of sum compositions. Sum compositions and sc-constraints are written in single quotes. Then there can be a WITH followed by certain options. The options can be:
- DBR is the double bound range of a sc-constrain. It is a 2-tuple stating the minimum and the maximum double bounds and rings which are allowed for a sum composition of this sc-constrain.
- CHG states the charge
If the fragment should be a neutral loss, this can be stated by setting the charge to zero with CHG = 0 or by writing AS NEUTRALLOSS after the sum composition or sc-constrain.
NOTE: The neutral loss is calculated always between the precursor mass and the fragment, never between two fragments.
examples
Define PC-O sc-constrains and PC-O's head group which is connected to the precursor mass:
DEFINE PR = 'C[30..48] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; DEFINE pcHead = 'C5 H15 O4 P1 N1' WITH CHG = 1;
Define PE sc-constrains and PE's head group which is connected to the precursor mass:
DEFINE PR = 'C[30..46] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; DEFINE peHead = 'C2 H8 O4 N1 P1' AS NEUTRALLOSS;
Define sc-constrains and fragments for PE-Plasmalogen:
DEFINE PR = 'C[30..46] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; DEFINE FRAG1 = 'C[14..26] H[20..80] O[3]' WITH DBR = (1.5,9), CHG = 1; DEFINE FRAG2 = 'C[14..26] H[20..80] N[1] O[4] P[1]' WITH DBR = (1.5,9), CHG = 1;
An arbitrary number of variables can be defined, but they are only valid for the current query. I.e. they are not valid in other queries of the same Run.
Part 2: The IDENTIFY section
The before defined variables are queried to the experiment database. The syntax is:
IDENTIFY <identification 1> AND <identification 2> AND ... <identification n>
The headline 'IDENTIFY' is followed by identifications which are connected by 'AND'. The result of an identification can be a singleton or a set, i.e. for some variables more than one mass is identified. This holds especially for sc-constraints. This section is the first filtering step. The section returns True if the boolean expression is true. The expression is true if the particular expressions are true:
An identification looks like this:
((<variable name> IN (MS1+/-|MS2+/-)+)?
Here does LipidXplorer check the existence of certain masses/fragment masses. The scope (level of MS) is stated after 'IN': The 'MS1+', 'MS1-', 'MS2+' and 'MS2-' tags point to the MS level where to look for the sum composition ('MS1+' means in positive MS, while 'MS2-' means in negative MS/MS).
Emulating (Multiple) Precursor Ion Scan / Neutral Loss Scan with MFQL
In the IDENTFIY section specify precursor ion scans (PIS) and neutral loss scans (NLS)can be defined. If the variable is a sc-constrain it emulates multiple PIS/NLS. Switching from PIS to NLS is done in the definition part. When a variable gets charge zero (CHG = 0) or the keyword AS NEUTRALLOSS is given then it is stated as neutral loss. Otherwise it is stated as (fragment) mass.
(Comment: The above feature should not be not mistaken with the LipidXplorer functionality to import PIS and NLS mass spectrometric acquisitions.)
Some examples:
# Phosphatedylcholine ether species DEFINE PR = 'C[30..48] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; DEFINE pcHead = 'C5 H15 O4 P1 N1' WITH CHG = 1; IDENTIFY # the MS mass should fit to 'PR' and it should have a MS/MS fragment mass fitting to 'pcHead' PR IN MS1+ AND pcHead in MS2+ ################################################################################ # Phosphatedylethanolamine DEFINE PR = 'C[30..46] H[30..200] N[1] O[8] P[1]' WITH DBR = (2.5,9), CHG = 1; DEFINE peHead = 'C2 H8 O4 N1 P1' WITH CHG = 0; IDENTIFY # marking PR IN MS1+ AND peHead in MS2+ ################################################################################ # PE Plasmalogen DEFINE PR = 'C[30..46] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1; DEFINE FRAG1 = 'C[14..26] H[20..80] O[3]' WITH DBR = (1.5,9), CHG = 1; DEFINE FRAG2 = 'C[14..26] H[20..80] N[1] O[4] P[1]' WITH DBR = (1.5,9), CHG = 1; IDENTIFY # marking PR IN MS1+ AND FRAG1 IN MS2+ AND FRAG2 IN MS2+
Part 3: The SUCHTHAT section
After the collection of specific masses, it is possible to add more constraints to the query. For example: the identification of PE Plasmalogen requires the marking of 'FRAG1' and 'FRAG2' which both contain several possibilities since they are sc-constraints (see example above) and a test if those two fragments in sum match the precursor mass, i.e. is "FRAG1 + FRAG2 == PR"? Such a constraint is formulated in the optional 'SUCHTHAT' section as boolean connected equations, unequations and functions. The syntax is:
SUCHTHAT (((NOT)? (<equation> | <unequation> | <function>)) | ((NOT)? (<equation> | <unequation> | <function>) (AND | OR))+) (WITH (<option> = <value>)+)?
The terms can be build up with the basic mathematical functions +, -, *, /. Parenthesis can also be used. The terms are connected as equations by '==' and as inequalities by '<', '>', '<=', '>=' and '!=' for not equal. The values for the terms can be marked masses (given with their variable name), floating point numbers or chemical sum compositions. Certain attributes of marked masses can be also addressed. This can be done by writing the attribute after the variable name connected with a dot. The intensity of the peak 'PR' for example is addressed as PR.intensity. A list of peak attributes can be found here: #List_of_peak_attributes
Functions
Additional to the attributes, SUCHTHAT supports the use of functions. The list of all functions can be found here: #List_of_functions
Part 4: The REPORT section
All successful identifications are piped to the REPORT section, where the format of the output is specified. In general the REPORT consists of a list of variables where each represents a column. The content of the variable is the content of the column. So is the following code generates a column with the name MASS and the m/z values of PR's identified species as content:
REPORT MASS = PR.mass
The next example reports the sum of the intensities of two fragments
REPORT INTENS = frag1.intensity + frag2.intensity
Mostly those fragments can be the same (so for example for 2 fatty acid scans), therefore LipidXplorer has a special function which does not sum intensities of same fragments:
REPORT INTENS = sumIntensity(frag1.intensity, frag2.intensity)
The syntax of REPORT is:
REPORT ((<variable name> = <variable> | <equation>)
The content of the variable can be any attribute and/or term as in the SUCHTHAT section. The REPORT section has an additional feature with which it is possible to generate lipid names or other formatted strings.
The syntax for this function is:
REPORT (<variable name> = "<format string>" % ((<list of variables for the format string>)+)
The string format works as follows: there are two strings to give which are separated with a %. The first string contains the output format, i.e. a string with placeholders. Placeholder can be: %d for decimal values, %.nf for floating point values with n decimals and %s for string values. The second string contains a list with the content of the placeholders according to their order. For example:
REPORT LIPIDNAME = "PC [%d:%d]" % (fa1PC.chemsc[C] + fa2PC.chemsc[C], fa1PC.chemsc[db] + fa2PC.chemsc[db])
The variable LIPIDNAME contains the string "PC [... : ...]". The first decimal value is filled with the sum of the carbon atoms of both fatty acids (fa1PC, fa2PC) and the second decimal value the sum of the double bonds. The output could be for example "PC [36:2]".
The format string variant is a Python gimmick, where MFQL uses standard Python commands. I.e. the format string is a python function (see here for more information).
Notes
- If a lipid was not found in a particular sample, its intensity is set to zero.
- If the isotopic correction corrects an intensity to zero or less than zero, it is set to '-1'
List of peak attributes
error
The difference between the theoretical mass (according to the sum composition) and the tagged mass from the spectrum. The error can be given in the 3 types:
- errppm -> error in ppm
- errda -> error in dalton
- errres -> error as resolution value
mass
The m/z value of the peak
chemsc
The chemical sum composition. For addressing certain elements of the sum composition, the element is to write in brackets after .chemsc. To get the number of C atoms from a formula for example:
PR.chemsc[C]
- frsc -> the chemical sum composition of the fragment. If the peak is defined as a (charged) fragment, it is the same as chemsc, if it is defined as a neutral loss, it returns the sum composition of the fragment.
- nlsc -> the chemical sum composition of the neutral loss. If the peak is defined as (uncharged) a neutral loss, it is the same as chemsc, if it is defined as a fragment, it returns the sum composition of the neutral loss of the precursor.
intensity
All the intensities of a mass from all the samples it occured. Note that intensity is mostly no single value but a list of intensities. One list entry for every sample the peak was found. If used in an equation or unequation, the whole list is considered. I.e. PR.intensity > 10000 is true if and only if all intensities are greater than 10000. It is possible to address only a part of all samples. This is done by writing the name of the sample group as string with wildcards (* and/or ?). E.g. is PR.intensity["*blanck*"] returning just the samples with the string blanck in their name. This could be all blanck samples. This feature allows to generate sample groups by naming the samples according to their group. So, a lot of different constraints can be stated, which increase the accuracy of the interpretation or even already interpret the result. E.g.
avg(PR.intensity["*blanck*"]) < avg(PR.intensity["*exp*"]) / 100
This statement asserts that the one percent of the average intensity of all experimental samples ("*exp*") should be greater than the average intensity found in the blanck sample. This simply throws out every "lipid", which is obviously noise.
binsize
The size of the bin of the peak coming from the averaging algorithm. The value is given in Dalton.
occ
Is the occupation of the peak. Occupation = nb. of occurences in the sample / nb. of samples
List of functions
isEven(n)
where n is an integer value. The function returns True, if n is even. E.g.: isEven(PR.chemsc[C]).
isOdd(n)
where n is an integer value. The function returns True, if n is odd.
avg(v.intensity)
where n is a variable. The function returns the average value of the intensities of n. E.g.:
avg(PR.intensity)
isStandard(v, scope)
where v is a variable and scope is "MS1+", "MS1-", "MS2+" or "MS2-". This function is special since it does not return anything. It enables the automatic calculation of standardizied intensities according to the given standard in v. I.e. Every intensity is calculated as relative to v.
sumIntensity(f1.intensity, f2.intensity, ...)
The function sumIntensity() is used for summing up intensities of different MS2 entries where multiple peaks are required for identification and quantification. In case of fragments with isotopic corrected place holders (see above)the following rules were implemented.
If all MasterScan entries in the MS2 for a particular molecule are place holders (i.e. all are set to '-1') then those values are just added and will result in <math>n_i\times -1</math> where ni is the number of the attributes.
If there is just one entry whose intensity is greater zero all − 1 place holders are threaded as zero and not added to the overall sum. In the presented example we assume that two entries in the MS2 where used for the sumIntensity() function:
F1 + F2 − > sumIntensity(F1.intensity,F2.intensity) − 1 + − 1 = − 2 0 + − 1 = − 1 1 + − 1 = 1 2 + − 1 = 2 2 + 0 = 2
That has following consequences when such results have to be interpreted:
A) intensity = 0 in this specific sample none of the required fragments was present
B) intensity < 0 in this sample some of the required fragments were found in the initial MasterScan but set '-1', none fragment above threshold (1) was present
C) intensity = -ni all fragments were below the threshold (1) after isotopic correction
D) intensity > 0 in this case at least one of the required fragments was after isotopic correction above the threshold (1)
Some examples
SUCHTHAT
# the number of 'C' atoms in 'PR's chemical sum composition should be odd
isOdd(PR.chemsc[C])
SUCHTHAT
# the sum of both fragments ('FRAG1', 'FRAG2') minus one 'H' should be equal to
# the precursor mass ('PR') and
# the intensity of 'FRAG2' should be bigger than 3/10th of the
# the intensity of 'FRAG1'
FRAG1 + FRAG2 - 'H1' == PR AND
FRAG1.intensity * 3 < FRAG2.intensity * 10
How LipidXplorer runs multiple MFQL queries
The principle of a LipidXplorer Run is the following: All queries run successively on the given MasterScan. For every query, LipidXplorer iterates through the list of MS masses of the MasterScan from smallest to the greatest and checks the conditions given in definition, IDENTIFY, SUCHTHAT and REPORT sections. I.e.
- it loads a MS mass
- it checks if it fits a given sum compostion or sc-constrain (definition and IDENTIFY section).
- it looks into its MS/MS spectrum (if provided) and does the same (definition and IDENTIFY section).
- the boolean constraints are checked (SUCHTHAT section) and if the result is
positive the MS mass is accepted and send to the REPORT section
Examples
Screen (without MS/MS experiments) for Phosphatidylcholine species
A "screen" is a fast identification based on only MS information. To do screening properly the masses should be high accurate, because otherwise the error of identification is too high.
The name of the query here is Phosphatidylcholine. Giving a name to a query is obligatory and has to be done for every query. We define the sc-constraint prPC (short for "precursor of PC") and state that it should be found in the positive MS spectra.
Names for variables are arbitrary. The user should try to give meaningful names in order to understand his query better.
The IDENTIFIY section urges LipidXplorer to look for the precursor mass into the MS spectrum.
In SUCHTHAT we use a function to restrict the result to lipids having an overall even number of carbon atoms. This means that the fatty acids of the lipid have to have both fatty acids even numbered or both odd numbered. Such, we can sort out lipids which we know they should not be in the organism we examine.
The REPORT section uses the following variables:
- 'MASS' returns the m/z value of the MS mass
- 'NAME' returns the lipid species' name, which consists of the number of carbon atoms and double bonds of the fatty acids. Those numbers we get from taking the number of carbons/double bonds from the sum composition (prPC.chemsc[C]/prPC.chemsc[db]) and reduce it by the carbons/double bonds belonging to the PC's head group and glycerol backbone.
- 'CHEMSC' returns the chemical sum composition
- 'INTENS' returns the abundance of the identified lipid species for all samples
- 'ERROR' returns the error of the finding in ppm.
########################################################## # Identify PC with checking the precursor mass # ########################################################## QUERYNAME = Phosphatidylcholine; DEFINE prPC = 'C[30..48] H[30..200] N[1] O[8] P[1]' WITH DBR = (2.5,9), CHG = 1; IDENTIFY # marking prPC IN MS1+ SUCHTHAT isEven(PC.chemsc[C]) REPORT MASS = prPC.mass; NAME = "PC [%d:%d]" % (prPC.chemsc[C] - 8, prPC.chemsc[db] - 5); CHEMSC = prPC.chemsc; INTENS = prPC.intensity; ERROR = "%2.2fppm" % (prPC.errppm); ; ################ end script ##################
The output of the query is the following:
This is a screen shot of spread sheet software holding the resulting data from the query. At the top are the variable names followed by the name of the query, then comes the content. Note, that for 'INTENS' the file name from which the sample data was taken is also written. Every entry in the result fulfills the constraints given in the query. If an expected value is not found then the query or the import settings should be refined.
Analysis of Phosphatidylcholine lipid species emulating PIS 184
Additionally to the former query we have a variable 'headPC' which contains the sum composition of the specific head group for PC which is found in the fragment spectra after MS/MS of a PC species. This variable is added as constraint in IDENTIFY. Thus a lipid is only identified if it fits to the constraints of prPC AND has a headPC fragment in its MS/MS spectrum. Again, we test the even numbers of carbons in SUCHTHAT, which ensure we do not find borderline masses, which actually cannot be in the sample. In the output we have additionally the abundance of the head group fragment with FRAGINTENS.
##########################################################
# Identify PCs with checking the precursor mass #
# AND check for PIS 184 in MS2 #
##########################################################
QUERYNAME = Phosphatidylcholine;
DEFINE prPC = 'C[30..48] H[30..200] N[1] O[8] P[1]' WITH DBR = (1.5,7.5), CHG = 1;
DEFINE headPC = 'C5 H15 O4 P1 N1' WITH CHG = 1;
IDENTIFY
# marking
prPC IN MS1+ AND
headPC in MS2+
SUCHTHAT
isEven(prPC.chemsc[C])
REPORT
MASS = prPC.mass;
NAME = "PC [%d:%d]" % ((prPC.chemsc - headPC.chemsc)[C] - 3, prPC.chemsc[db] - 1.5);
CHEMSC = prPC.chemsc;
ERROR = "%2.2fppm" % (prPC.errppm);
INTENS = prPC.intensity;
FRAGINTENS = headPC.intensity;;
################ end script ##################
Application of Boolean operation "AND" for identification of PE-plasmalogen
An example for a whole script:
###########################################################
##### find PE-plasmalogens with MS2 in positive mode ######
###########################################################
# define sf-constrains and fragments for PE-Plasmalogen
DEFINE PR = 'C[30..46] H[30..200] N[1] O[7] P[1]' WITH DBR = (1.5,8), CHG = 1;
DEFINE FRAG1 = 'C[14..26] H[20..80] O[3]' WITH DBR = (1.5,9), CHG = 1;
DEFINE FRAG2 = 'C[14..26] H[20..80] N[1] O[4] P[1]' WITH DBR = (1.5,9), CHG = 1;
IDENTIFY
# marking
PR IN MS1+ AND
FRAG1 IN MS2+ AND
FRAG2 IN MS2+
SUCHTHAT
# the sum of both fragments ('FRAG1', 'FRAG2') minus one 'H' should be equal to
# the precurosor mass ('PR') and
# the intensity of 'FRAG2' should be bigger than 3/10th of the
# the intensity of 'FRAG1'
FRAG1 + FRAG2 - 'H1' == PR AND
FRAG1.intensity * 3 < FRAG2.intensity * 10
REPORT
# first column is the precursor mass
MASS = PR.mass,
# second is the lipids name generated with Python's string formatting function
NAME = "PE-O [%d:%dp / %d:%d]" % (FRAG1.frsc[C], FRAG1.frsc[db] - 2, FRAG2.frsc[C], FRAG2.frsc[db] - 2),
# third is the precursor's chemical sum composition
CHEMSC = PR.chemsc,
# forth the intensity
INTENS = PR.intensity,
# fifth the sum of the error of both fragments in ppm
ERROR = FRAG1.errppm + FRAG2.errppm;;
More Examples
More examples can be found in the MFQL collection provided in the LipidXplorer wiki.