Difference between revisions of "LXMerge"
(→Usage of LXMerge) |
(→Usage of LXMerge) |
||
| Line 19: | Line 19: | ||
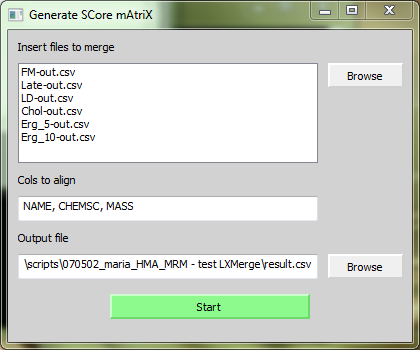
[[Image:screenshot1.PNG|center|LXMerge]] | [[Image:screenshot1.PNG|center|LXMerge]] | ||
| − | |||
# Click on Browse to select or Drag'n Drop your result files. | # Click on Browse to select or Drag'n Drop your result files. | ||
| Line 31: | Line 30: | ||
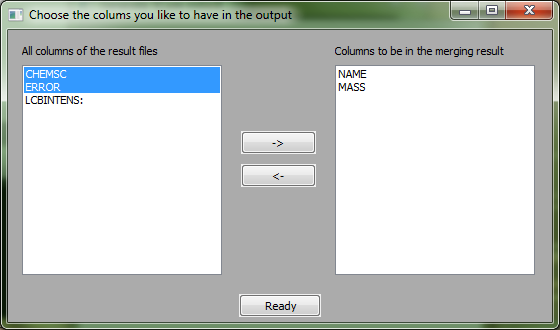
[[Image:screenshot2.PNG|center|LXMerge]] | [[Image:screenshot2.PNG|center|LXMerge]] | ||
| − | |||
The left side contains all columns which were found in the resulting files. All columns having a ':' at the end are samples. The two buttons '->' and '<-' will move the column names to the end of the right or respectively left list. The order of the chosen columns will be remained. | The left side contains all columns which were found in the resulting files. All columns having a ':' at the end are samples. The two buttons '->' and '<-' will move the column names to the end of the right or respectively left list. The order of the chosen columns will be remained. | ||
Revision as of 17:11, 8 October 2010
Contents
Purpose of LXMerge
LXMerge aligns and merges result files coming from LipidXplorer. The columns to be aligned are assigned by the user. Also, the output columns as well as their order are assigned by the user.
Use case one: using specific occupation threshold settings
Often you have samples groups coming from different sources, for example from different phenotypes of a particular organism. Every phenotype is analysed including biogical and technical repeats leading to groups of samples assigned to each phenotype, respectively.
To have an accurate lipid identfication the occupation threshold option from the import settings of LipidXplorer can be used to assert that a peak is present in a given percentage of samples. The problem is, that there could be lipids which are not present in some samples. An occupation threshold setting for all samples would give an incomplete result.
With LXMerge one can overcome this issue. For this use case it would make sense to import every sample group in an own MasterScan with a stringend occupation threshold setting. Then use LXMerge to align the results by an unique key, which is a column with enties which do not repeat. The result at the end is a merged file containing the aligned results.
Use case two: align results from different machines
When you used different machines and therefore different MasterScan for the same sample, LXMerge makes it easy to align and compare all the results.
Usage of LXMerge
- Click on Browse to select or Drag'n Drop your result files.
- Write the name of the columns you want to align. Separate each with a semicolon.
- Click Browse to select your output file.
- Click on 'Start'
The left side contains all columns which were found in the resulting files. All columns having a ':' at the end are samples. The two buttons '->' and '<-' will move the column names to the end of the right or respectively left list. The order of the chosen columns will be remained.
- Click on '->' to move a selected column to the right side
- Click on '<-' to move a selected column back